Ruby T.S. Lam, John M. Collins, Alexander B. Smetana, Saju Nettikadan, NanoInk Inc.
March 15, 2011 — The ability to place individual cells at defined locations and control their microenvironment has numerous applications in the field of cell biology. Cells respond to their microenvironment and the resulting extracellular signals [1, 2]. The ability to control the cellular microenvironment enables investigation of biochemical and topological cues on various cell behavior, such as cell adhesion, differentiation, and molecular signaling pathways.
Cell morphology has been shown to be closely related to their function at an individual cell level as well as at the tissue and organ level [3]. The long rod shape of myocytes, the elongated shape of neurons, and the spindle shape of fibroblasts are all believed to play a role in the function of their respective tissues. For in vitro systems, these morphologies are influenced by the topographical elements and the geometry of the adhesion pads [4]. Furthermore, biochemical signals from the surface and intercellular signaling from neighboring, interacting cells are critical factors in mediating cell behavior [5]. Controlling the cellular microenvironment is particularly interesting for stem cell studies: commitment of stem cells to different specific lineages has been demonstrated to be dependent on cell shapes that are induced by topography and biochemical cues [6]. Further, the ability to place cells at defined positions on a substrate is critical to the development of cell-based sensing and cell-based drug discovery.
Microcontact printing is currently the most common method for micropatterning cells [7]. This method is used to deposit biomaterials or other chemical entities at defined locations on a surface using a stamp made of a soft material such as Polydimethylsiloxane (PDMS). The limitations of microcontact printing are that not all proteins can be reliably deposited, patterning of multiple materials is also difficult to achieve [8]. The need for a mask and stamp for each pattern makes this method very rigid and not amenable to rapid changes in pattern definition. The surfaces are generally fabricated through silicon microfabrication methods which are not biologically friendly, are expensive, and do not suit to rapid changes in pattern design.
The nanolithography platform, NLP 2000, was designed to provide a simple solution to achieve high precision placement of nano- to micro-sized features with nanoscale precision. The process of deposition of material is based on Dip Pen Nanolithography (DPN), an established method of nanofabrication in which materials are deposited onto a surface using a sharp tip [9, 10]. The tool is capable of patterning a wide range of materials with feature sizes from 50nm to 10µm over an area of 40 x 40mm. The features can be placed with nanoscale precision using a three-axis closed-loop stage.
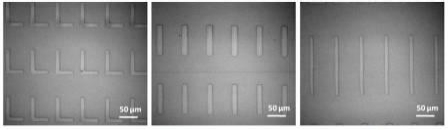 |
|
Figure 1. Acrylic polymer patterns printed on glass substrates by NLP 2000. |
The NLP 2000 has been used to deposit proteins, DNA, lipids, polymers, hydrogels, silanes and thiols on many different substrates at subcellular scales. Printing these various materials allows the design and construction of microenvironments for cell-based studies. In this report, we present results demonstrating the capability of the technology to construct topographical and biochemical features at subcellular scales and show the cellular response to these features.
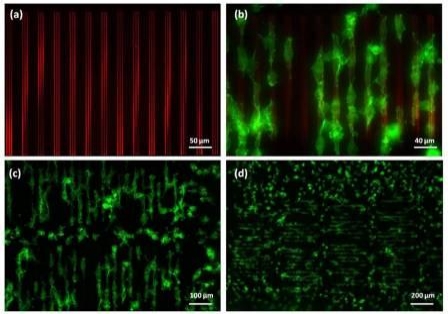 |
|
Figure 2. Fluorescence images of (a) patterned acrylic polymer-rhodamine topography; (b) 3T3 cells selectively adhered to and aligned along the direction of the polymer topography; (c) & (d) elongated cells covered 70% of the patterned area (over 1.5 x 0.7mm) at 1.5hrs after cell seeding. Green – actin filaments; red – acrylic polymer patterns. |
Influencing cell behavior using topographical features is an attractive strategy for investigating many aspects of cellular function. Features at subcellular dimensions are particularly interesting because it closely mimics the physical features of the in vivo microenvironment. An ultraviolet (UV) curable polymer was used to successfully fabricate topographies on a glass surface. Despite the high ink viscosity (>20,000 cps), the researchers directly deposited it onto a glass surface at subcellular scales with nanoscale precision. Polymer lines that are as fine as 500nm in width and 20nm in height were achieved. Figure 1 shows the bright field images of several polymer patterns constructed on the same chip. These structures were shown to favor cell adhesion. NIH/3T3 fibroblasts (25,000 cells/cm2) were cultured on the substrate in low serum media. Cell attachment was observed on approximately 70% of the patterned features within two hours of cell seeding. Cell morphology was altered due to the patterned polymer on the glass substrate. Figure 2 shows the cellular shapes resulting from the printed polymer topographies that covered a 1.5 x 0.7mm area of the glass substrate. Most cells attached to the topography were selectively aligned on top of the ridges and elongated along the direction of the grating. It is known that focal adhesion dynamics are dictated by topography. Here, spacing between the ridges played an important role in modulating cell spreading and morphology. In this case, cells residing on ridge structures smaller than 2µm showed only minimal focal contact, whereas more pronounced cell elongation and spreading was observed on topographies between 10 to 15µm in total width. In addition, the greatest alignment was observed on grating spaced 20µm apart, and declined as the width decreased. Cells on non-patterned regions also attached to the substrate, but were mostly rounded. On flat surfaces coated with the polymer, the cells bound spread out randomly, demonstrating that the chemical characteristics of the acrylic polymer did not contribute to the elongated morphology. The molecular mechanisms responsible for topography-mediated responses still remain unclear. The data demonstrates that the tool is a flexible platform for construction of topographically patterned substrates for cell biological studies.
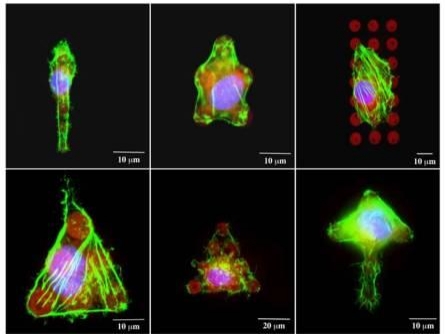 |
|
Figure 3. Single 3T3 fibroblasts attached to fibronectin patterns. Cells conform to the shape of the pattern provided. Green – actin filaments; blue – nucleus; red – fibronectin |
Another field of interest for cell patterning studies is the cellular response to surface bound biomolecular signals. DPN can be used to fabricate subcellular-scale ligand features for cell studies. The tool enables the user to produce substrates with precise positioning of individual protein ligands, allowing one to control the spacing between ligands with nanometer precision. Here, we demonstrate the technique to directly deposit extracellular matrix proteins onto functionalized glass surfaces. Given the constructive nature of Dip Pen Nanolithography, printing was carried out at ambient conditions, which avoided any protein denaturation due the harsh environmental conditions. A series of fibronectin dot arrays (8μm in diameter and 15μm pitch) were designed and fabricated. The surface was then treated with 2% BSA to prevent nonspecific cell adhesion. NIH/3T3 fibroblasts were added at high density (100,000 cells/cm2) for 30 minutes, at which point non-adherent cells were washed and removed. Cells that remained attached to the fibronectin spots adopted the orientation of the micropatterns. Figure 3 shows fluorescence images of single 3T3 fibroblasts patterned into various shapes using fibronectin. Controlling the shape and location of cells distributed on a patterned substrate at a multicellular scale opens the door to investigate the effects of cell-cell contact in a well-controlled fashion.
Conclusion
Several earlier studies have demonstrated that both topography and biochemical cues are important in regulating cellular behavior. We have demonstrated that Dip Pen Nanolithography, as a fabrication tool, mimics cellular microenvironments in both approaches. It is likely that a truly novel biomaterial scaffold mimicking the in vivo conditions will consist of both topographical and biochemical cues. This nanolithography process can integrate these cues and enable tailor-making of biomimetic microenvironments from subcellular scales to multicell levels.
Acknowledgments
Dip Pen Nanolithography and DPN are registered trademarks of NanoInk Inc.
Ruby T.S. Lam received her PhD/Cambridge University and is an Applications Scientist at NanoInk Inc., 8025 Lamon Avenue, Skokie, IL 60077 USA; ph.: 8477453634; [email protected]
John M. Collins received his PhD/University of Illinois at Chicago and is an Applications Scientist at NanoInk Inc.
Alexander B. Smetana received his PhD/Kansas State University and is an Applications Scientist at NanoInk Inc.
Saju Nettikadan received his PhD/ The Ohio State University and is the director of applications development at NanoInk Inc.
References
1. Shen CJ, Fu JP, Chen CS. Patterning Cell and Tissue Function. Cellular and Molecular Bioengineering, 2008, 1, 15-23.
2. Kulangara K, Leong KW. Substrate topography shapes cell function. Soft Matter, 2009, 5, 4072-4076.
3. Dalby MJ, Childs S, Riehle MO, Johnstone HJH, Affrossman S, Curtis ASG. Fibroblast reaction to island topography: changes in cytoskeleton and morphology with time. Biomaterials, 2003, 24, 927-935.
4. Biggs MJP, Richards RG, Dalby MJ. Nanotopographical modification: a regulator of cellular function through focal adhesions. Nanomed.:Nanotechno., Biol. Med., 2010, 6, 619-633.
5. Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol., 2005, 23, 47-55.
6. Yim EKF, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res., 2007, 313, 1820-1829.
7. Altomare L, Riehle M, Gadegaard N, Tanzi M, Fare S. Microcontact printing of fibronectin on a biodegradable polymeric surface for skeletal muscle cell orientation. Int. J. of Art. Organs, 2010, 33, 535-543.
8. Ganesan R, Kratz K, Lendlein A. Multicomponent protein patterning of material surfaces. J. of Mater. Chem., 2010, 20, 7322-7331.
9. Piner RD, Zhu J, Xu F, Hong SH, Mirkin CA. "Dip-pen" nanolithography. Science, 1999, 283, 661-663.
10. Sekula S, Fuchs J, Weg-Remers S, Nagel P, Schuppler S, Fragala J, Theilacker N, Franueb M, Wingren C, Ellmark P, Borrebaeck CAK, Mirkin CA, Fuchs H, Lenhert S. Multiplexed Lipid Dip-Pen Nanolithography on Subcellular Scales for the Templating of Functional Proteins and Cell Culture. Small, 2008, 4, 1785-1793.
Follow Small Times on Twitter.com by clicking www.twitter.com/smalltimes. Or join our Facebook group

